- Search NCIBI Data
(e.g. diabetes, csf1r) - Login

 About Our Tools and Data
About Our Tools and Data
About Our Data
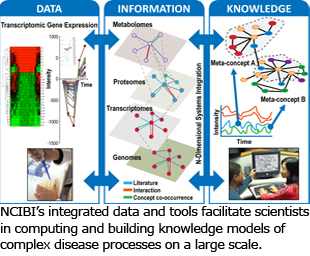
We integrate and make publicly available experimental datasets, data from numerous biomedical and protein interaction databases, and PubMed/Medline. Our data also include information we derive from mining text through powerful natural language processing (BioNLP) and statistics for genes and metabolites significantly enriched for a wide range of concepts (e.g. Gene Ontology annotations, Medical Subject Headings (MeSH). We provide access to various database resources through Application Programming Interfaces (API) and on request.
Specifically, NCIBI databases include:
NCIBI GENE database: Gene data integrated from public sources.
NCIBI HUMDB database: Metabolomics data from public and internal sources. Integrates Reactions, Compounds, Genes, and Proteins.
NCIBI MiMI database: Protein, Gene, Pathway and Interaction data integrated from over 10, e.g. HPRD and KEGG. The database is rebuilt on a regular basis.
Natural Language Processing (NLP) techniques in PubMed and PubMed Central databases: NLP in these databases tags gene names, gene name synonyms, and metabolites from segmented sentences. Protein-protein interactions and their annotations are also extracted. Techniques include sentence segmentation, named entity tagging, and parsing sentences into trees. From trees, the NLP determines phrase structures within sentences and grammatical relationships between pairs of words.
NCIBI Pubmed database: All source files from the National Library of Medicine's (NLM) Pubmed database, downloaded nightly and updated to the database combined with BioNLP data.
NCIBI Pubmed Central database: All full-text biomedical articles from the Pubmed Central Open Access (PMCOA) database, with source files downloaded monthly and updated to the database combined with BioNLP
About Our Tools
Tools draw on our diverse databases. The best starting point for those unfamiliar with NCIBI tools is the "Try Our Tools" page. Links to video tutorials and accompanying documentation are also available on this page.
NCIBI provides the software architecture and tools to search our data and graphic and tabular workspaces for exploring the data and generating new knowledge about complex biomedical problems at a molecular and systems level. The key objective is to give scientists access to data that will help them gain novel insights into conceptual relationships.
NCIBI tools and resources are available free of charge to the research community for internal use. Please refer to the General Terms of Use for details. Additional terms of use may apply to individual tools. For more information, consult the website of the tool in question.
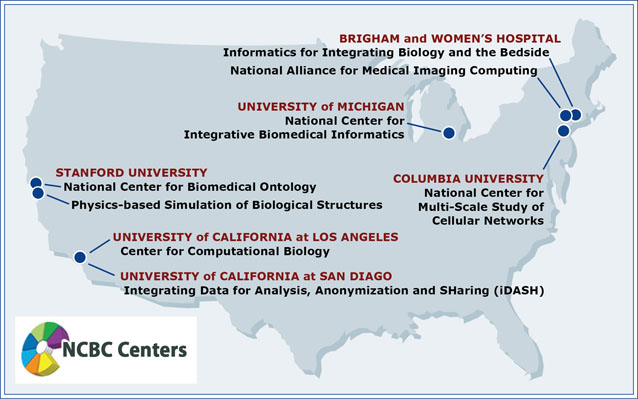
Searching with Biositemaps Across National Centers
NCIBI tools, as well as tools developed by affiliated collaborators and other National Centers for Biomedical Computing, are all catalogued into a Biositemaps directory. Those familiar with NCIBI resources can go directly to the tool they want to use through the Biositemaps search page, which can also be accessed through links on Try our Tools.
By querying with Biositemaps you can search for tools by tool name, tool type/category, affiliation, or other criteria. Tools can be accessed directly from the search results page.

 NCIBI on Facebook
NCIBI on Facebook NCIBI RSS Feed
NCIBI RSS Feed

