- Search NCIBI Data
(e.g. diabetes, csf1r) - Login

Tools & technology seminar series
Seminar information is now listed on the
U-M Department of Computational Medicine and Bioinformatics website.
Thursdays
12 noon - 1:00p.m. EST
Room 2036 Palmer Commons
Ann Arbor, MI
HighLights
The National Center for Integrative Biomedical Informatics (NCIBI) is one of eight National Centers for Biomedical Computing (NCBC) within the NIH Roadmap. The NCBC program is focused on building a universal computing infrastructure designed to speed progress in biomedical research. NCIBI was founded in September 2005 and is based at the University of Michigan as part of the Department of Computational Medicine and Bioinformatics (DCMB).
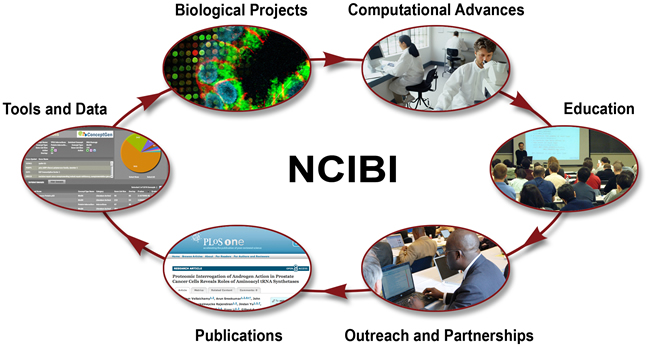
Led jointly by teams of computational and biomedical scientists, NCIBI:
Integrates vast amounts of diverse, multi-scale data and derived knowledge, including context-appropriate molecular biology information from emerging experimental data; gene, protein, and metabolite databases; and the published literature.
Collaborates to determine how these data sets can best be represented and developed into resources that will advance research and facilitate biomedical discoveries.
Creates relevant tools for analytically exploring the data to uncover and validate functional associations and possible causal and conditional relationships involved in mechanisms of complex physiological processes or diseases.
Develops an array of tutorials, seminars, documentation, and other training materials to assure both the usability and usefulness of NCIBI tools.
Disseminates discoveries and processes to biomedical communities worldwide through publications, presentations, national partnerships and collaborators, and e-networking initiatives such as an RSS feed and the NCIBI gateway.
News
The tranSMART Foundation Wins Awards at the BioIT World 2015 Conference!
Dr. Gilbert S. Omenn discusses the emerging HUPO Human Proteome Project on Australian SkyNews.
New Tools and Services
We are pleased to announce a release of Metscape 2.2. The most prominent changes include a more versatile workflow and enhanced visualization features.
Gene2MeSH and Metab2MeSH versions 3.0 have been released.
RECENT Publications


 NCIBI on Facebook
NCIBI on Facebook NCIBI RSS Feed
NCIBI RSS Feed

